Effects of Tumor Microenvironmental Factors on DNA Methylation and Radiation Sensitivity in A549 Human Lung Adenocarcinoma
Article information
Abstract
Background
Tumor response to anticancer therapies can much be influenced by microenvironmental factors. In this study, we determined the effect of these microenvironmental factors on DNA methylation using A549 human lung adenocarcinoma cell line.
Materials and Methods
We subjected A549 cells to various conditions mimicking tumor microenvironment including hypoxia, acidosis (sodium lactate), oxidative stress (H2O2), bystander effect (supernatant from doxorubicin (Dox)-treated or irradiated cells), and immune cell infiltration (supernatant from THP-1 or Jurkat T cells). Genomic DNA was isolated from these cells and analyzed for DNA methylation. Clonogenic cell survival, gene expression, and metabolism were analyzed in cells treated with some of these conditions.
Results and Discussion
We found that DNA methylation level was significantly decreased in A549 cells treated with conditioned media from Dox-treated cells or Jurkat T cells, or sodium lactate, indicating an active transcription. To determine whether the decreased DNA methylation affects radiation sensitivity, we exposed cells to these conditions followed by 6 Gy irradiation and found that cell survival was significantly increased by sodium lactate while it was decreased by conditioned media from Dox-treated cells. We further observed that cells treated with conditioned media from Dox-treated cells exhibited significant changes in expression of genes including BAX and FAS (involved in apoptosis), NADPH dehydrogenase (mitochondria), EGFR (cellular survival) and RAD51 (DNA damage repair) while sodium lactate increased cellular metabolism rather than changing the gene expression.
Conclusion
Our results suggest that various tumor microenvironmental factors can differentially influence DNA methylation and hence radiosensitivity and gene expression in A549 cancer cells.
Introduction
Tumor microenvironment is composed of various populations of cells including not only cancer cells but also endothelial cells, pericytes, fibroblasts, and immune cells interacting closely with each other leading to create a unique environment such as hypoxia and acidosis. It is now well established that tumor microenvironment can drastically modulate the tumor sensitivity to anticancer therapies either directly or indirectly. For example, tumor hypoxia is known to be a direct resistance factor for ionizing radiation because the lack of oxygen prevents formation of cytotoxic DNA radicals, the ultimate goal of radiotherapy to produce cell killing effects in tumors [1]. Pericytes and fibroblasts have been shown to indirectly exert resistance to anticancer therapies by increasing survival signals in endothelial cells [2] or in cancer cells [3] by secreting growth factors for these cells such as vascular endothelial growth factor and fibroblast growth factor. We have previously demonstrated that bone marrow-derived myeloid cells, those give rise to monocytes and tumor-associated macrophages could also contribute to the tumor recurrence after radiotherapy by expressing extracellular matrix degrading enzymes such as matrix metalloproteinase-9 [4].
Recently, radiosensitivity of tumors has been demonstrated not only influenced by inherited gene expression but also epigenetic modification such as DNA methylation [5]. DNA methylation is mediated by DNA methyltransferase (DNMTs) transferring methyl group at the 5′ position of the cytosine base in DNA, leading to hypermethylation in CpG islands, which are present in 70% of all mammalian promoters [6]. Hypermethylation has been frequently observed in lung cancers particularly in those genes including APC, CDH1, CDH3, CDKN2A/p14(ARF), CDKN2A/p16, MGMT [7]. A recent study has interestingly demonstrated that GABBR2 gene is hypermethylated in EGFR-mutated lung adenocarcinoma and that erlotinib treatment causes downregulation of not only GABBR2 but also DNMTs, mediating the drug-induced apoptosis [8]. Previously, we and others have reported that DNMT inhibition by using chemicals including 5-azacytidine, 5-aza-2′-deoxycytodine, zebularine, and psammaplin A [9] or genetic knockdown approaches [10] could potentially radiosensitize A549 lung adenocarcinoma cells via inhibition of DNA repair.
In the present study, we have investigated effects of tumor microenvironmental factors on DNA methylation in A549 cells and its influence on radiosensitivity. We originally hypothesized that these tumor micronenvironmental factors may increase DNA methylation in cancer cells thus providing a potential rationale to combine a DNMT inhibitor with radiotherapy. However, we found in this study that some of microenvironmental factors including sodium lactate and conditioned media obtained from doxorubicin-treated cells or Jurkat T cells resulted in actually decreased DNA methylation although these factors differentially influenced gene expression and radiosensitivity, indicating a complex epigenetic modulation in tumors within the tumor microenvironment.
Materials and Methods
1. Cell culture and treatments
A549 human lung carcinoma cells were maintained in Dulbecco’s Modified Eagle Medium (Welgene, Gyeongsan, Korea) supplemented with 10% Fetal Bovine Serum (Corning, Corning, NY). Cells were exposed to a tumor microenvironmental factor, which was adopted from studies published by other investigators with minor modifications outlined in Table 1. Briefly, cells were treated with 300 μM H2O2 (Daejung, Siheung, Korea) for 24 hours; 10 mM sodium L-lactate (Sigma-Aldrich, St. Louis, MO) for 24 hours; fresh conditioned media harvested from A549 cells that had been pre-treated with 10 μM doxorubicin (Sigma-Aldrich, St. Louis, MO) for 24 hours at 24 hours prior to the harvest; fresh conditioned media harvested from 20 Gy irradiated A549 cells at 24 hours prior; or fresh supernatant harvested from confluent THP-1 cells or Jurkat T cells at 24 hours prior. Conditioned media was filtered using 0.22 μM filter (ADVANTEC, Dublin, CA) prior to the exposure to fresh A549 cells.
2. Cell viability
Cell viability was examined by trypan blue staining where single cells were collected and mixed with 0.4% trypan blue (Gibco, ThermoFisher, Waltham, MA) at 1:1 ratio. Cells were examined in a hemocytometer using a bright-field microscope (Zeiss, Oberkochen, Germany).
3. Methylated DNA Quantification
DNA from A549 cells was isolated by using Accuprep Genomic DNA Extraction kit (Bioneer, Daejeon, Korea) and the DNA concentration was measured using Nanodrop (Nanodrop 2000, ThermoFisher, Waltham, MA). Total DNA methylation status was quantified by using Methylated DNA Quantification Kit (Abcam, Cambridge, UK) according to the manufacturer’s instructions.
4. Ionizing radiation and cell surviving fraction
Cells were treated as above with conditioned media obtained from Dox-treated cells; THP-1 cells; or 10 mM sodium lactate 24 hours prior to 6 Gy ionizing radiation, which was done using 137Cs irradiator with the dose rate 3.41 Gy·min−1 (Gammacell 3000 Elan, Nordion, Canada). After irradiation, cells were plated for 2 weeks to measure surviving fractions. Colonies were stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO) in 20% methanol (Daejung, Siheung, Korea).
5. RNA extraction and cDNA synthesis
Total RNA was isolated by using TRIzol reagent (Ambion Inc., ThermoFisher, Waltham, MA) according to the manufacturer’s instructions. Concentrations of RNA samples were measured by Nanodrop, and 2 μg of RNA was mixed with Superasin (Invitrogen, Carlsbad, CA), DNase (Promega, Madison, WI), and RNase free H2O for 15 minutes at room temperature, followed by addition of 10 mM EDTA (Bioneer, Daejeon, Korea) for 8 minutes at 65°C. After then, 10 mM dNTP (Bioneer, Daejeon, Korea) and Random Primer (150 μg·mL−1) (Invitrogen, Carlsbad, CA) were added. Samples were then heated for 30 seconds at 95°C, 5 minutes at 65°C, 1 minute at 4°C. DTT (0.1 mol·L−1) (Sigma-Aldrich, St. Louis, MO), superasin, and MgCl2 (25 mM) (Promega, Madison, WI) were then added for 2 minutes at 37°C, followed by addition of Reverse Transcriptase (Promega, Madison, WI). The reaction mixture was incubated for 10 minutes at 25°C, 120 minutes at 37°C, 15 minutes at 75°C, and then cooled at 4°C.
6. Quantitative real time-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed with StepOne (ThermoFisher) using SYBER Green reagents (Promega, Madison, WI). qPCR primers for genes are as follows: β -actin Fwd (5′ GGACTTCGAAGAGATGG 3′), β -actin Rev (5′ AGCACTGTGTTGGCGTACAG 3′), HK2 (Hexokinase2) Fwd (5′ TCTATGCCATC-CCTGAGGAC 3′), HK2 Rev (5′ TGGACTTGAATCCCTTGGTC 3′), GLUT-1 (Glucose transporter-1) Fwd (5′ CTTCACTGTCGTGTCGCTGT 3′), GLUT-1 Rev (5′ CCAGGACCCA-CTTCAAAGAA 3′), LDHA (Lactate dehydrogenase A) Fwd (5′ GAGGTTCACAAGCAGGTGGT 3′), LDHA Rev (5′ CCCA-AAATGCAAGGAACACT 3′), MCT-1 (Monocarboxylate transporter-1) Fwd (5′ TGACCATTGTGGAATGCTGT 3′), MCT-1 Rev (5′ CCTTTTTCTGCTCGTTTGCT 3′), MCT-4 (Monocarboxylate transporter-4) Fwd (5′ TGGGATGGGACTGAC-TTTTC 3′), MCT-4 Rev (5′ CCATGTGCAGACAAACTGCT 3′), ND-1 (NADH dehydrogenase subunit-1) Fwd (5′ CTACTACAACCCTTCGCTGAC 3′), ND-1 Rev (5′ GGATTGA-GTAAACGGCTAGGC 3′), SDH (Succinate dehydrogenase) Fwd (5′ GGATCTTGTTCCCGATTTGA 3′), SDH Rev (5′ ATTTGTC-TCCGTTCCACCAG 3′), CYB- α (Cytochrome b-245 alpha chain) Fwd (5′ CGCTTCACCCAGTGGTACTT 3′), CYB- α Rev (5′ GAGAGCAGGAGATGCAGGAC 3′), COX-2 (Cytochrome c oxidase subunit-2) Fwd (5′ CCCTTACCATCAAATCAATTGGCC 3′), COX-2 Rev (5′ ATTGTCAACGTCAA-GGAGTCGC 3′), PERK (Protein kinase R-like endoplasmic reticulum kinase) Fwd (5′ AGGGGCACTCCTTTGAACTT 3′), PERK Rev (5′ ATTTGCTACTGGTGGGCTTG 3′), ATF-6 (Activating transcription factor-6) Fwd (5′ CAGCGAATAGCCCAGTGAAT 3′). ATF-6 Rev (5′ TTCTGAGAGGGCAGCCTTTA 3′), GADD-34 (Protein phosphatase 1 regulatory subunit 15A) Fwd (5′ AAA-CCAGCAGTTCCC-TTCCT 3′), GADD-34 Rev (5′ CTCTT-CCTCGGCTTTCTCCT 3′), XBP-1s (X-box binding protein 1) Fwd (5′ TCACCCCTCCAGAACATCTC 3′), XBP-1s Rev (5′ AC-AGAGAAAGGGAGGCTGGT 3′), GRP-78 (Glucose regulated protein-78) Fwd (5′ GGTGAAAGACCCCTGACAAA 3′), GRP-78 Rev (5′ GTCAGGCGATTCTGGTCATT 3′), GRP-94 (Glucose regulated protein-94) Fwd (5′ TGCCAAGGAAGGAGTGAAGT 3′), GRP-94 Rev (5′ GTTGCCAGACCATCCGTACT 3′), BAX (BCL2 associated X, apoptosis regulator) Fwd (5′ TTTGCTTCAGGG-TTTCATCC 3′), BAX Rev (5′ CAGTTGAAGTTGCCGTCAGA 3′), FAS (Fas cell surface death receptor) Fwd (5′ CAAGGGATTGGAATTGAGGA 3′), FAS Rev (5′ TGGAA-GAAAAATGGGCTTTG 3′), Cyclin D Fwd (5′ AACTACCTGGACCGCTTCCT 3′), Cyclin D Rev (5′ CCACTTGAGCTTGTTCACCA 3′), CDK2 (Cyclin dependent kinase 2) Fwd (5′ AAATTCATGGATGCCTCTGC 3′), CDK2 Rev (5′ CAGGGA-CTCCAAAAGCTCTG 3′), EGFR (Epidermal growth factor receptor) Fwd (5′ CAGCGCTACCTTGTCATTCA 3′), EGFR Rev (5′ AGCTTTGCAGCCCATTTCTA 3′) BRCA (BRCA, DNA repair associated) Fwd (5′ GGTGGTACATGCACAGTTGC 3′), BRCA Rev (5′ ACTCTGGGGCTCTGTCTTCA 3′), RAD51 (RAS associated with diabetes protein 51) Fwd (5′ TCACGGTTAGAGCAGTGTGG 3′), RAD51 Rev (5′ GATCACGCTATTGCACTCCA 3′), PARP1 (Poly(ADP-ribose) polymerase 1) Fwd (5′ GCTCCTGAACAATGCAGACA 3′), PARP1 Rev (5′ TCCTGATGATCTCGGCTTCT 3′), CHK2 (Checkpoint kinase 2) Fwd (5′ CTGTTGGGACTGCTGGGTAT 3′), CHK2 Rev (5′ CGTAAAACGTGCCTTTGGAT 3′). PCR steps were: 2 minutes at 50°C and 10 minutes at 95°C, after then, 15 seconds at 95°C and 1 minute at 60°C for 40 cycles. The relative quantification of mRNA was analyzed by using the comparative temperature (Ct) method, as the manufacturer’s protocol.
7. Seahorse cellular metabolism measurements
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured by a Seahorse XFe96 analyzer (Seahorse Biosciences, Agilent, North Billerica, MA). Cells were plated in a 96-well plate and treated with growth media containing 10 mM sodium lactate or conditioned media harvested from Dox-treated cells. Mitochondrial metabolism was evaluated with the XF Cell Mito Stress Kit (Agilent, Santa Clara, CA) provided with 1 μM oligomycin, 1 μM cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin-A. Glycolysis was measured using 10 mM glucose (Gibco, ThermoFisher, Waltham, MA), 1 μM oligomycin (Sigma-Aldrich, St. Louis, MO), and 50 mM 2-deoxyglucose (2-DG) (Sigma-Aldrich, St. Louis, MO).
8. Statistics
Statistical comparisons were performed by student’s t test or one-way ANOVA using Prism software version 4.00 (GraphPad Inc., La Jolla, CA). p-values<0.05 were considered to be statistically significant.
Results
1. Effects of tumor microenvironmental factors on DNA methylation in A549 cells
To determine an effect of tumor microenvironment on DNA methylation in cancer cells, we introduced various microenvironmental factors such as hypoxia, lactate, oxidative stress, immune cell infiltration, and chemotherapy/radiotherapy-treated conditions (Table 1) to A549 cells. We observed that neither hypoxia (Figure 1A) nor H2O2-induced oxidative stress (Figure 1C) resulted in any significant changes in DNA methylation. Sodium lactate, on the other hand, caused a significant decrease in DNA methylation levels (Figure 1E). Sodium lactate at this concentration did not lead to any significant changes in pH of the culture media (Figure 1G). Cells at these conditions maintained at least 95% viability as determined by trypan blue (Figure 1B, 1D, and 1F).
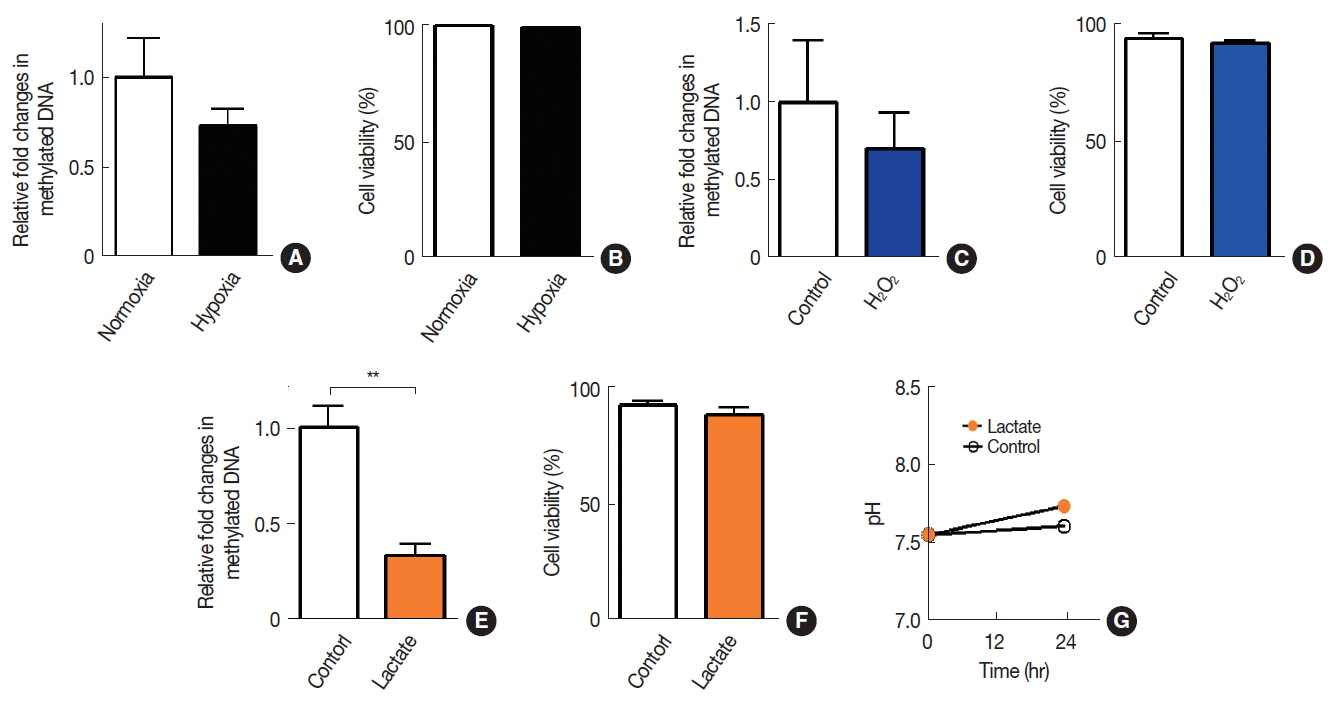
Effects of hypoxia, oxidative stress, or acidosis on DNA methylation in A549 cells. DNA methylation (A, C, and E) and cell viabilities (B, D, and F) were quantified in A549 cells exposed to hypoxia (A and B), H2O2 (C and D), or sodium lactate (E and F). (G) Extracellular pH measurement in media added with vehicle control (control) or lactate. Data are the mean±s.e.m. for n≥3 per group. **indicates p<0.01, determined by Student’s t test.
To introduce ‘immune cell infiltration’ component of the tumor microenvironment, we obtained conditioned media from THP-1 myeloid cells or Jurkat T cells and exposed these conditioned media to A549 cells. We observed that there was a significant decrease in DNA methylation by the exposure of conditioned media obtained from Jurkat T cells but not from THP-1 cells (Figure 2A). We further introduced fresh conditioned media obtained from A549 cells that had been treated with doxorubicin (Dox) for 24 hours prior to the conditioned media harvest and observed that this conditioned media significantly decreased DNA methylation in A549 (Figure 2B). Cell viability at this condition, however, exhibited an approximately 20 % decrease, indicating a significant bystander cytotoxicity (Figure 2C). To investigate whether the bystander effect would result in a decrease in DNA methylation, we harvested conditioned media from irradiated A549 cells and exposed it to freshly cultured A549 cells. We observed that conditioned media harvested from irradiated cells did not lead to a significant change in DNA methylation (Figure 2D) while it caused a significant decrease in cell viability (Figure 2E).
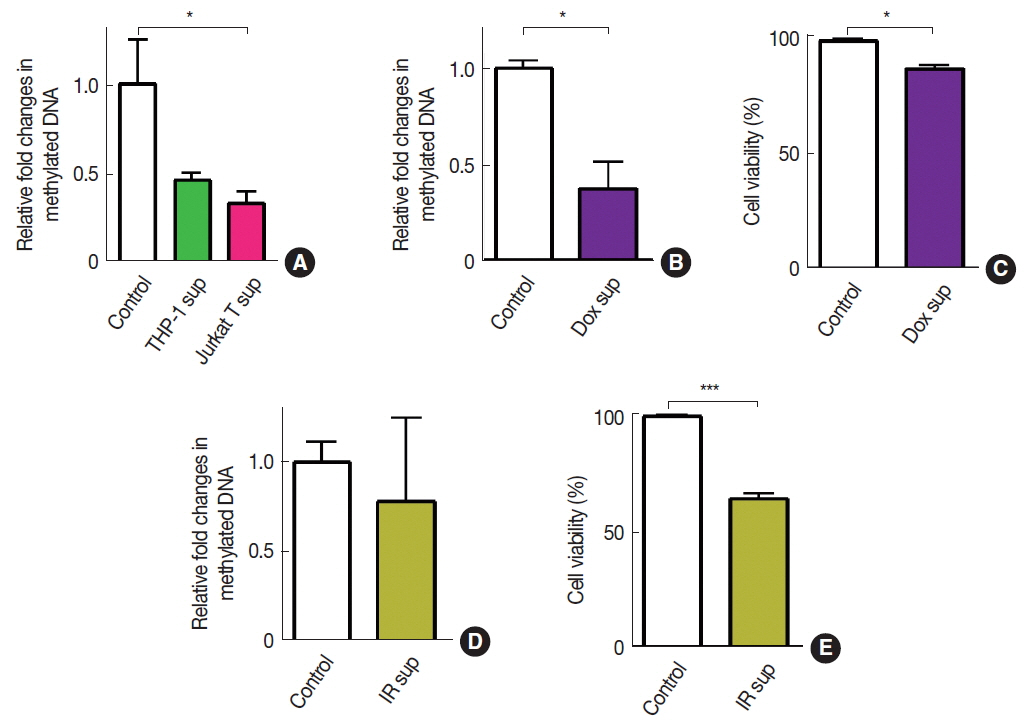
Effects of immune cell infiltration or bystander effects on DNA methylation in A549 cells. DNA methylation (A, B, and D) and cell viabilities (C and E) of A549 cells treated with conditioned media obtained from THP-1 or Jurkat T cells (A), Dox-treated cells (B and C), or 20 Gy irradiated cells (D and E). Data indicate the mean±s.e.m. for n≥3 per group. * and *** indicates p<0.05 and 0.001, respectively, as determined by Student’s t test.
2. Effects of DNA methylation on radiosensitivity of A549 cells
Decreased DNA methylation may indicate increased transcriptional activities [11], which in turn may affect the radiosensitivity of cells. To test this hypothesis, we exposed those microenvironmental factors above, which resulted in decreased DNA methylation to A549 cancer cells (i.e., sodium lactate, conditioned media harvested from cells treated with Dox, or conditioned media from Jurkat T cells) and irradiated them to 6 Gy to determine whether the decrease in DNA methylation would lead to a survival advantage. By performing clonogenic cell survival assay, we observed that 6 Gy irradiation resulted in approximately 95% cell kill (Figure 3A). We further found that sodium lactate treatment significantly increased the cell survival (Figure 3A) while conditioned media obtained from Dox-treated cells significantly decreased the survival (Figure 3A) compared to the vehicle-treated control, all of which were irradiated with 6 Gy. Exposure of conditioned media obtained from Jurkat-T cells did not cause any significant changes in the survival compared to the control (Figure 3A).
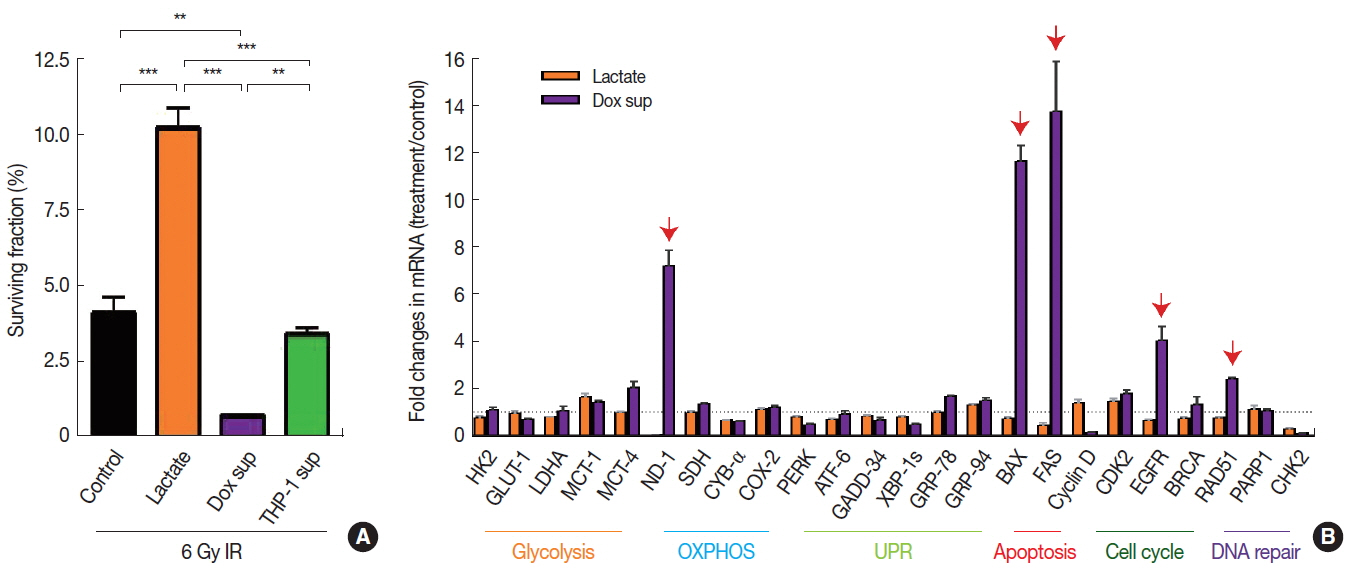
Radiosensitivity and gene expression changes in A549 cells treated with lactate or conditioned media obtained from Dox-treated cells. (A) Surviving fraction of 6 Gy irradiated cells as expressed as percentage relative to unirradiated control for each group. Cells were treated for 24 hr as indicated, prior to 6 Gy irradiation. Data are the mean±standard deviation for duplicate samples. (B) Gene expression changes as indicated by fold changes in mRNA levels in treated cells versus untreated cells. Data are the mean±s.e.m. for triplicate determinations. Red arrows indicate significantly increased gene expressions (fold changes greater than 2). ** and *** indicate p<0.01 and 0.001, respectively, determined by one-way ANOVA.
3. Dox-treated conditioned media increase gene expression pathways involving apoptosis and DNA repair while lactate modulates cellular metabolism to affect radiosensitivity
Next we examined which gene transcripts were up- or down-regulated affecting the radiosensitivity of A549 cells treated with sodium lactate or conditioned media obtained from Dox-treated cells by performing quantitative real-time polymerase chain reaction. We found in A549 cells treated with conditioned media obtained from Dox-treated cells that the expression of several genes including BAX and FAS (involved in apoptosis), NADPH dehydrogenase (ND-1; mitochondria), EGFR (cellular survival) and RAD51 (DNA damage repair) were significantly increased (Figure 3B). In contrast, we did not find any significant changes in the gene expression in cells treated with sodium lactate (Figure 3B).
Lactate has recently been reported to use as a fuel source in the nearby respiring cancer cells by being taken up and converted back to pyruvate, a major energy substrate for mitochondrial oxidative phosphorylation [12]. We hypothesized that lactate may modulate cellular metabolism in order to increase radiation resistance that we observed above (Figure 3A). To test our hypothesis, we measured oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) to measure OXPHOS and glycolysis, respectively in A549 cells treated with lactate. We further included a group where A549 cells were treated with conditioned media obtained from Dox-treated cells. We found that sodium lactate but not conditioned media from Dox-treated cells significantly increased basal OCR (Figure 4A) and basal respiration (Figure 4B) in A549 cells, consistent with our hypothesis that lactate may promote OXPHOS. There was no significant change in the maximal respiration ability in cells treated with either lactate- or conditioned media obtained from Dox compared to vehicle control (Figure 4C). Upon measuring ECAR, we also observed that sodium lactate increased both glycolysis level and glycolytic capacity (Figure 4D–F). Conditioned media obtained from Dox-treated cells also lead to an increase in glycolysis (Figures 4D and 4E) while it has no effect on glycolytic capacity (Figure 4F). The results therefore suggest that lactate promotes OXPHOS cellular metabolism increasing radioresistance while conditioned media obtained from Dox-treated cells increases gene expression involved in apoptosis and DNA repair to increase radiosensitivity of A549 cells.
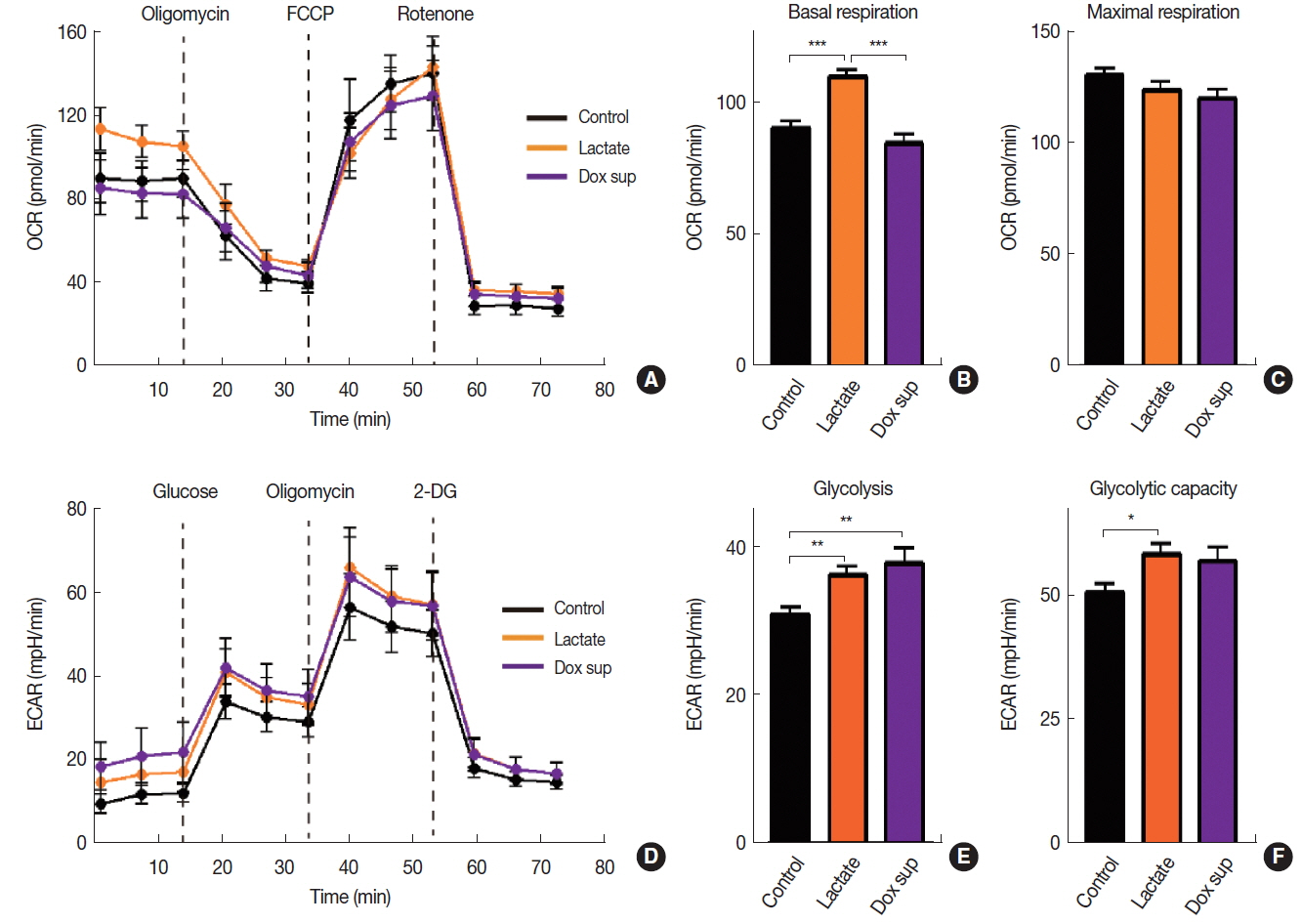
Cellular metabolism of A549 cells treated with lactate or conditioned media obtained from Dox-treated cells. Oxygen consumption rate (OCR, A) and its quantifications as basal respiration (B) and maximal respiration capacity (C). Extracellular acidification rate (ECAR, D) and its quantification as glycolysis (D) and glycolytic capacity (E). Data are the mean±s.e.m. for n≥10 per group. *, **, and *** indicate p<0.05, 0.01 and 0.001, respectively, determined by one-way ANOVA.
Discussion
In this study, we aimed to determine whether the tumor microenvironment alters DNA methylation thereby influencing radiosensitivity of A549 human lung adenocarcinoma cells. We originally hypothesized that microenvironmental factors would increase DNA methylation, hence a combination with DNA methyltransferase inhibitor would sensitize tumors to ionizing radiation. However, we found that some (including lactate and conditioned media obtained from Jurkat T cells or Dox-treated cells) but not all microenvironmental factors that we tested decreased, not increased the global DNA methylation status in A549 cells. Consistent with this, Mathot and colleagues [13] have also found that cancer-associated fibroblasts co-cultured with breast human cancer cells did not lead to a significant DNA methylation changes. Tumor-associated fibroblasts isolated from non-small cell lung cancer patients on the other hand have revealed promoter hypermethylation-associated SMAD3 silencing, leading to exacerbated TGF-β signaling [14]. It is likely that tumor cells or stromal cells exposed to tumor microenvironment may turn on gene expression machinery in order to adapt and survive in such harsh conditions and that this active transcription may be manifested as epigenetic changes including hypomethylation in DNA or acetylation in histone. Supporting our hypothesis, a study has demonstrated that bystander effects resulting from 20 Gy irradiated cranium in rats have led to decreased DNA methylation in long interspersed nucleotide element-1 (LINE-1) gene, which then gave rise to an increased expression of microRNA-194 to downregulate DNA methyltrasferase-3a and methyl-binding protein 2 in the unirradiated spleen [15]. It is interesting to note that Puthli and colleagues [16] have reported similar findings to ours where they observed that 5 μg·mL−1 Dox-induced bystander effect resulted in an increased radiation sensitivity in HeLa cells. They further demonstrated that Dox-induced bystander effect caused increased nuclear P53 and γ-H2AX protein expression in cells. These are also in good agreement with our results where we observed increased expression of apoptosis-involved genes including BAX and FAS (Figure 3B) and DNA repair gene RAD51 (Figure 3B) because BAX [17] and FAS [18] are known to be downstream targets of TP53. Moreover, it has been demonstrated that RAD51 has been shown to be recruited to γ-H2AX foci after DNA damage [19].
Recent studies have interestingly indicated that the use of DNMT inhibitors may lead to a significant epigenetic modification in stromal cells including immune cells. Zhang and colleagues [20] have detected DNA methylation signature especially in PD-1 in peripheral blood monocytes and T cells during the progression of hepatocellular carcinoma, indicating a possible synergistic antitumor activity between DNMT inhibitor and immune checkpoint blockades. Furthermore, the use of 5-azacytidine has demonstrated in cytokine-treated cancer-associated fibroblasts that their proinvasive properties acquired by hypermethylation of SHP-1 leading to activation of STAT3 signaling can be effectively inhibited by DNMT inhibitor [21]. It is worthy to note that once cytokine-induced epigenetic repression is established, targeting only DNMT1 but no other isoforms of DNMT was able to inhibit the epigenetic reprogramming of cytokine-induced cancer-associated fibroblasts [21], suggesting that DNMT isoform may differ in regulating the gene expression depending not only on the type of cells but also spatiotemporal epigenetic modification.
We also observed that not all these parameters affected radiosensitivity nor gene expression changes. We found while lactate increased, conditioned media obtained from Dox-treated cells decreased the cell survival after 6 Gy irradiation. Gene expression analyses through candidate approach using qRT-PCR revealed that there were a number of genes were upregulated in cells treated with conditioned media obtained from Dox-treated cells, including BAX and FAS (involved in apoptosis), NADPH dehydrogenase (ND-1; mitochondria), EGFR (cellular survival) and RAD51 (DNA damage repair). It is reasonable to hypothesize that cells treated with this conditions may trigger cell survival signal partly through DNA damage repair while those experiencing bystander effect through toxic metabolites in the conditioned media may trigger apoptotic genes such as BAX and FAS. Upregulation of ND-1 is interesting although it has been previously reported that ND-1 expression is upregulated in the embryonic mouse fibroblasts exposed with doxorubicin as a protective/cellular defense mechanism [22]. Lactate treatment, on the other hand, did not cause any significant changes in the gene expression. We reasoned that lactate may stimulate cellular metabolism via oxidative phosphorylation as suggested previously by Sonveaux and colleagues [12], instead of modulating the expression of genes that we examined. Sonveaux and colleagues [12] have demonstrated that exogenous treatment of sodium lactate at a concentration of 10 mM to SiHa cells enforced oxidative metabolism, as measured by electron paramagnetic resonance. Indeed, we found that this identical treatment to A549 cells resulted in a significant increase in the basal oxygen consumption ratio, an indicator of oxidative phosphorylation, which may have increased cellular survival following 6 Gy irradiation.
Collectively, our data demonstrate that some of tumor microenvironmental factors in A549 cells lead to a decrease in global DNA methylation, some of which exhibited the gene expression changes while others modulated cellular metabolism. These suggest that there may be a complex mechanism exist between the tumor microenvironment and cancer cells, modulating epigenetic modification hence gene expression in a reciprocal manner.
Acknowledgements
This study was supported by grants from the National Research Foundation, Ministry of Science and Information, Communication, and Technology (ICT; grant no. NRF-2018-R1A2B6004453 and NRF-2017M2A2A7A01015968 to G.O.A; NRF-2016R1D1A1B03934691 and NRF-2017M2A2A7A 01070925 to H.J.K) and the Ministry of education (BK21 Plus, 10Z20130012243), Korea. Y.E.K (NRF-2012H1A-2A1002871) and B.J.H. (NRF-2013H1A2A1032808) are Global PhD fellowship recipients from the Ministry of Science, and ICT.
